
SARS-CoV-2 ORF9c responsible for immune evasion and efficient infection of host
The current COVID-19 pandemic comprises a full spectrum of cases ranging from critically severe or terminally ill patients to asymptomatic cases, which make up the vast majority. In more severe cases, immune dysregulation is thought to play a key role. A recent study by researchers at the Sanford Burnham Prebys Medical Discovery Institute and the University of California San Francisco and published on the preprint server bioRxiv* in August 2020 shows that a protein encoded by the subgenomic sequence ORF9c of SARS-CoV-2 is responsible for the low antiviral response in infected humans, enabling the successful replication of the virus in the host. This could lead to successful inhibition of viral immune evasion mechanisms.
Bioinformatics Analysis of ORF9c
The study used proteomics, interactome and transcriptional analysis passed through bioinformatics tools, to examine the effects of this small, 73-residue protein. Prior research has shown that such viral evasion mechanisms are a prime target for therapeutic intervention, to allow the host immune system to clear the virus more effectively. Most current vaccine candidates are directed at inhibiting viral entry via the spike protein, while remdesivir, an approved anti-SARS-CoV-2 drug, targets viral replication and assembly.
The current study looks at the accessory viral protein ORF9c, which has been shown by earlier proteomic and interactome studies to affect the way the host cell operates through many cellular signaling pathways. These include immune signaling via interferons (IFNs) and interleukins (ILs). This protein maintains the structure of organelles involved in viral replication as well as the virus itself, and the researchers show here that its sole expression alters cellular networks such that the outcome closely simulates full-fledged viral infection.
ORF9c Encodes Unstable Transmembrane Protein
The ORF9c protein is a very unstable protein, shared with SARS-CoV, which indicates it plays a role in causing the disease. Phylogeny and sequence alignment studies show that different coronaviruses have mutant variants of the protein, with the closest being in the bat coronavirus ORF14.
The analysis of the protein predicts that it has a transmembrane sequence at the C-terminal region, unlike SARS-CoV. The reading frame can extend by three amino acids with a single nucleotide mutation. This makes SARS-CoV-2 ORF9c the only such protein in a human coronavirus to contain a transmembrane domain. This ability to anchor itself to the host cell could increase its virulence and pathogenicity, by helping it to interact with the IFN pathway, changing antigen presentation, and immune evasion capabilities.
The researchers looked at the role of this protein in an epithelial lung cancer cell line. It interacts with multiple membrane-associated proteins scattered in many cellular compartments, as might be expected from the presence of the transmembrane domain, including those systems that involve protein synthesis and transport like the endoplasmic reticulum, Golgi apparatus, and mitochondria.
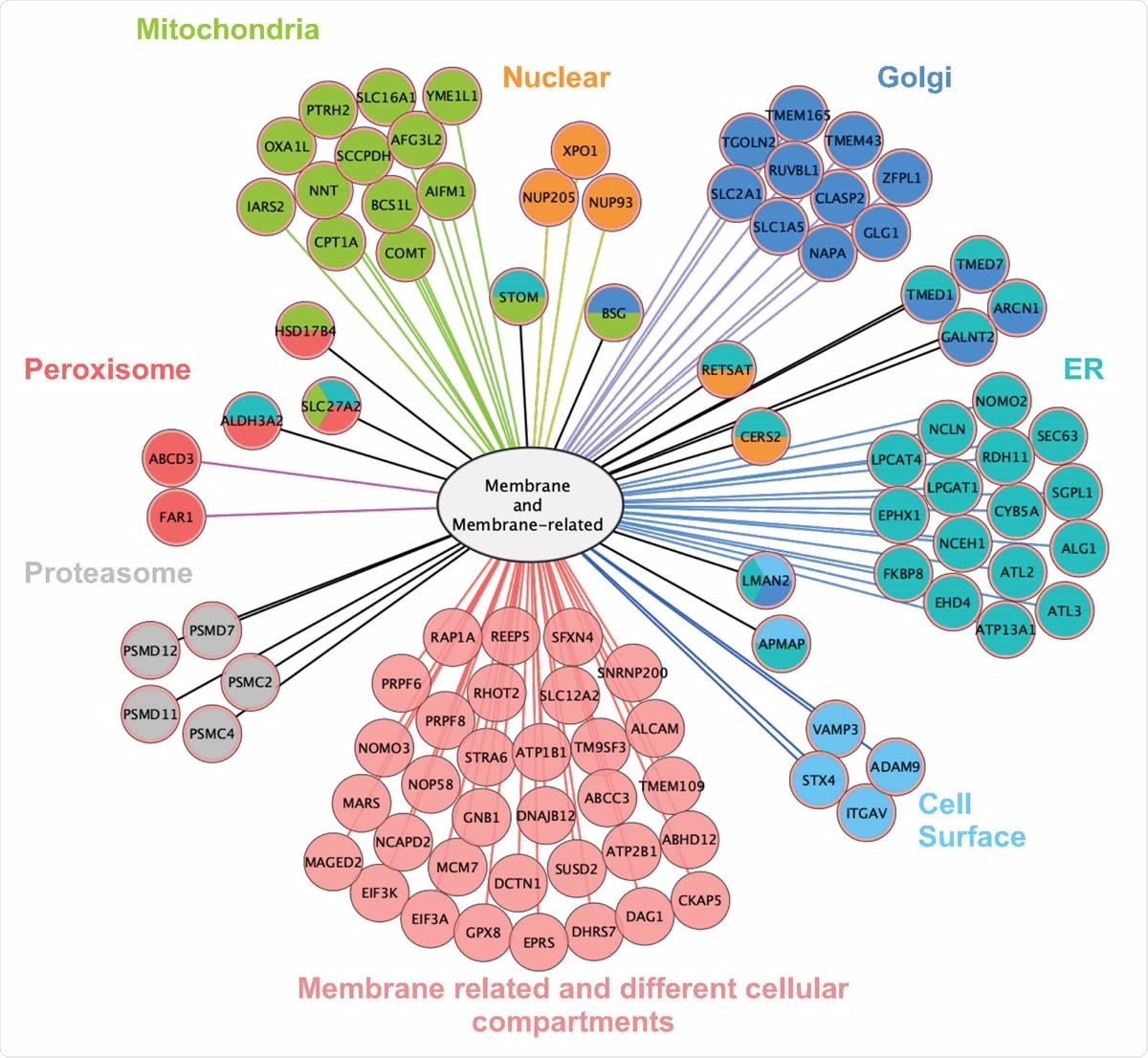
Interactome of ORF9c is based on LC-MS/MS of ORF9c-interacting proteins immunoprecipitated from A549 24 h after transfection. Left: Number of ORF9Cinteracting proteins according to their cell compartment (from Gene Ontology). Total values exceed 100% because some proteins are assigned / located in more than one compartment. Right: Protein subcellular localization map for the portion of the ORF9c interactome that is either a membrane protein or a membrane-related protein (as defined by Gene Ontology).
ORFc Proteomics Shows Downregulation of Immune Signaling
The researchers found that different proteins were expressed in the transfected and untransfected samples, with the significant change being protein downregulation in 144 cases, and upregulation in 14 cases. The most significant change was in IFN signaling, antigen presentation, and innate immune pathways, including pattern recognition receptors. Proteins in the MAPK/ERK2 pathway were increased. They also found that small concentrations of ORF9c can induce changes in cellular pathways, inhibiting IFN, immune recognition, and ubiquitin components at the protein level, contributing to immune evasion.
ORF9c Reduces Transcription of Immune Signaling Pathways
The researchers went on to assess transcriptional changes due to ORF9c expression. They found a higher number of transcripts to be differentially regulated than proteins, but the pattern was similar in that most involved immune signaling. However, the specific pathways were different in many cases.
The biggest changes were on the complement and some inflammatory pathways, such as antigen presentation and immune signaling. However, some unique transcriptional changes were in the induction of IL-6 and p38 MAPK signaling pathways, not mirrored in proteomic analysis.
Common ORF9c-Related Changes in Both Analyses
The researchers found that while both pathways showed the same pathways underwent changes in the same direction, the number of components affected in each pathway differed at protein and transcript level. An analysis of the interactions among the various components showed that downregulation was mainly observed concerning transcripts and proteins related to chemotaxis, complement, interferon, and antigen presentation.
Stress-related pathways and histone acetylation transcripts were induced by ORF9c, perhaps because transcriptional regression mediates some of the ORF9c-produced changes in gene expression.
Proteasome Inhibitors Reverse ORF9c Effects in Part
The addition of a proteasome inhibitor caused downregulation of some transcripts and proteins, but not all. The most significant change was with the ubiquitin pathway (UBP) components, and the unfolded protein response (UPR), which is vital for the regulation of cell cycle pathways. The researchers postulate that this may mean that these pathways take part in the breakdown of ORF9c. Again, it may mean that the ability to reduce important cell signaling pathways required for antiviral defenses depends on the presence of VCP.
The use of a different cell line may account for the differences between the findings of the current study and earlier research, which used HEK293 cells, as well as other criteria. The changes caused by exposure to this single protein are recapitulated in cells infected by the intact virus.
The greatest changes were in the loss of regulation of the IFN system, along with cytokines involved in the TNF and STAT signaling and innate immune factors. IFN signaling is involved in mounting an antiviral response, and in many of the disease features found in COVID-19, which can thus be traced back to the activity of this protein.
Implications
Less than 2% of patients have a detrimental mutation in the transmembrane domain encoding sequence. This will need further study to understand how this affects clinical outcomes.
The link to histone acetylation may indicate a mechanism for transcriptional repression caused by this protein, including the upregulation of stress-induced genes following exposure to this viral protein. This could restrict immune signaling and promote immune evasion.
The UBP association also harmonizes with that of cellular immune component downregulation, indicating that perhaps the former changes mediate the stability of other proteins, which in turn causes reduced cytokine signaling and decreased innate immune responses.
UPR components also destabilize the ORF9c protein, causing it to be recognized as a misfolded protein by the host cell. This protein binds to the UPR and the UBP, resulting in higher proteasome activity. They found that inhibitors of VCP, which mediates the engagement of the protein with the UPR, reduces the transcriptional repression effects of ORF9c on components of the immune system. Proteasomal inhibitors also have similar but less consistent effects.
These findings indicate new avenues for therapeutic drugs to target viral virulence and disease features. A means of preventing the membrane anchoring role of this protein could also be key to preventing and treating immune evasion caused by SARS-CoV-2. Further studies are needed to validate these findings in animal models.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Andres, A. D. et al. (2020). SARS-CoV-2 ORF9c Is a Membrane-Associated Protein that Suppresses Antiviral Responses in Cells. bioRxiv preprint. doi: https://doi.org/10.1101/2020.08.18.256776. https://www.biorxiv.org/content/10.1101/2020.08.18.256776v1

































No hay comentarios:
Publicar un comentario